Research Article - (2025) Volume 4, Issue 2
Screening and Identification of tsRNA Expression Profiles in Adriamycin-Resistant Acute Myeloid Leukemia Cells
Received Date: Mar 03, 2025 / Accepted Date: Apr 02, 2025 / Published Date: Apr 10, 2025
Copyright: ©©2025 Fuxue Meng, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Meng, F., Li, L., Jia, W., Yang, X., Xie, X. (2025). Screening and Identification of tsRNA Expression Profiles in Adriamycin-Resistant Acute Myeloid Leukemia Cells. J Surg Care 4(2), 01-08.
Abstract
Background: Refractory and relapse, caused by resistance to chemotherapy, which is the primary treatment for acute myeloid leukemia (AML), are the major hindrance in the cure of AML patients. Currently, tRNA-derived small RNAs (tsRNAs) are considered to be novel and potential non-coding RNAs that are involved in various cellular processes and play an important role in cancer progression. However, the expression of tsRNAs in AML resistant cells remain unclear.
Methods: In this study, the expression profiles of tsRNAs in AML resistant cells were determined by arraystar small RNA microarray analysis, further detailed analysis and expression verification were carried out.
Results: Our study reveals the distribution of tsRNA in AML resistant cells and confirms that expression of tsRNAs including up- regulated genes tRF5-GlyCCC and 5'tiRNA-GlnCTG, and 5'tiRNA-ArgTCG and tRF5-TyrGTA down-regulated genes in AML resistant cells. We further performed GO and KEGG analyses on the identified genes to better comprehend their molecular mechanisms.
Conclusions: This study clarified the tsRNA expression profiles of AML drug-resistant cells and verified the differential tsRNA expression, suggesting that tRF5-GlyCCC and 5'tiRNA-GlnCTG may be biomarkers of poor prognosis in AML. Nevertheless, 5'tiRNA- ArgTCG and tRF5-TyrGTA may actively regulate AML to counter chemotherapy resistance. This provides a new insights for the mechanism and therapeutic targets of chemoresistance in AML.
Keywords
Acute Myeloid Leukemia, Chemoresistance, tsRNAs, Biomarkers
Abbreviations
AML: Acute myeloid leukemia ADM: Adriamycin
HL-60/ADM Cells: HL-60 Adriamycin-resistant cells APL: Acute promyelocytic leukemia
CLL: Chronic lymphoid leukemia ANG: Angiogenin
SncRNAs: small non-coding RNAs tRNAs: transfer RNAs
tsRNAs: tRNA-derived small RNAs tRFs: tRNA-derived fragments
tiRNAs: tRNA-derived stress-induced RNAs GO: Gene Ontology
KEGG: Kyoto Encyclopedia of Genes and Genomes
Introduction
Acute myeloid leukemia (AML) is a molecularly and cytogenetically heterogeneous hematological malignancy characterized by epigenetic changes of genetic molecules in myeloid progenitor cells, which accounts for about 80% of adult acute leukemia [1,2]. New therapeutic drugs have been exploited to improve the overall survival of patients as the in-depth understanding of the pathogenesis and resistance mechanism [3]. However, some high-risk karyotype AML patients develop sign chemotherapy failure. Outside of acute promyelocytic leukemia (APL), the 5-year survival rate of adult patients with AML remains poor (less than 40%) [4]. Most patients experienced short or long- term relapse due to acquired secondary resistance after complete remission, resulting in the vast majority of AML patients still dying from this complication based on different relapse/refractory mechanisms [5]. Dynamic and heterogeneous genes expression make up a complex chemotherapy-resistant interaction network, leading to frequent occurrence of refractory and recurrent AML and poor prognosis.
Mounting studies have suggested that small non-coding RNAs (sncRNAs) are important regulators in a variety of biological processes, including cell differentiation and cellular stress responses [6]. Improved sequencing platforms and methods allowing us to perceive that mature transfer RNAs (tRNAs) can also generate sncRNA molecules through site-specific cleavage of various RNases such as angiogenin (ANG) and dicer [7]. tRNA- derived small RNAs (tsRNAs), involved in cell processes such as proliferation, metastasis, and apoptosis [8-10]. The diagnostic potential of tsRNAs have also been reported in prostate cancer, liver cancer, and renal cell carcinoma [11,12]. Although the function of tsRNAs in cancers are still not completely clear, promissing as the tumor biomarkers [13]. tsRNAs have also been shown to be indispensable in the early and progressive stages of certain types of cancer, including B-cell lymphoma, chronic lymphocytic leukemia (CLL), and lung cancer [14-17].
There is accumulating evidence that tsRNAs are dysregulated in various cancers, suggesting that tsRNAs may play an important role in cancer development and progression [18]. However, there are rare studies on the tsRNA expression profiles of AML resistant cells, especially no in adriamycin-induced HL-60 resistance. Xia comprehensively characterized the expression pattern of circulating sncRNAs in blood and bone marrow and their alteration signature between healthy controls and AML patients [19]. What's more, the team further demonstrates the important role of METTL1/ WDR4 in AML leukaemogenesis, which provides a promising target candidate for AML therapy [20]. In this case, the research on the drug resistance function of tsRNA is still in the early stage. This study explored and provided the expression of tsRNA in HL- 60/ADM resistant cells for the first time,.hoping to provide new insights into chemotherapy resistance and for the discovery of new prognostic markers in AML.
Method
Cell Culture, Drug Resistance Induction and Detection
HL-60, human promyelocytic leukemia cells, purchased from Kunming cell bank, KCB2014051YJ. After receiving the cells, we strictly follow the instructions to culture and induce drug resistance. HL-60 cells(untreated HL-60 cells) and HL-60/ ADM(Adriamycin-resistant cells) were incubated in RPMI 1640 medium containing 10% FBS. The initial induction concentration of adriamycin (ADM) in HL-ADM cell was 0.1 µg/mL, and culturing was continued until HL-60 cells grew and proliferated normally at 2 µg/mL ADM; HL-60 cells were collected and counted, and cultured for 24 h. ADM was added with differences in concentrations of 10 µL and cells were incubated for 24 h, then 10 µL of CCK-8 solution was added and further incubated in the incubator for 4 h and detectd. Extracted total proteins concentration determined.
SDS-PAGE electrophoresis was performed to tested the expreesion of MDR1 protein. In brief, cells were collected by centrifugation, and total protein was obtained after lysis with RIPA total protein lysis buffer (aiqiandu, Ba1004). The protein concentration of the samples was determined using a BCA protein concentration determination kit. SDS-PAGE rophoresis was performed, including sample processing, gel preparation and sample loading, membrane transfer, antibody incubation, and detection of target proteins by chemilumines. After membrane transfer, blocking solution was added and incubated at room temperature for 1 h; the blocking solution was removed, and the diluted primary antibodies β-in (1:6000), MDR1 (1:1000) (Both from San Ying Biological) were added; incubated overnight 4â??; the diluted primary antibodies were recovered, and washed three times with TBST each for 5 min; the diluted secondary antibodies were added, incub at room temperature for 30 min, and washed four times with TBST on a shaker at room temperature, each for 5 min. ECL mixed (Baiqiandu, Ba1059) was dropped on the protein side of the membrane to detect protein bands. ImageJ was used to analyze gray value of the target bands (Part of the methods refer to Li's reported [21].
Arraystar Small RNA Microarray Analysis
Arraystar small RNA microarray analysis was performed comply with manual operation procedure. In brief, RNA samples quantity and integrity were detected by QC’d and gel electrophoresis respectively. The labeled RNA was hybridized onto Arraystar Human small RNA Microarray and the array was scanned. After the array image was collected, the data is analyzed and normalized. Differential expression of small RNAs between the groups was identified by folding change (FC) and statistical significance (P-value) thresholds. Hierarchical clustering heat maps, scatter maps and volcano maps were drawn to show the expression patterns of small RNA.
Characteristics of Differentially Expressed Genes
We performed characteristic analysis in the total differentially expressed genes of miRNA, pre-miRNA, tRNA and tsRNA by Excel 2019. Moreover, in order to better understand the possible functions of different types of tsRNA, we made a Schematic diagram of derivation of different types of tsRNAs.
Functions of Differentially Expressed-Related tsRNAs
In the purpose of understand the molecular functions of differentially expressed tsRNAs, categorization statistics were performed base on the reported studies, tsRNAs that associated piRNA-like functions; Up-regulated and down-rugulated tsRNAs which associated with the devolepment of chronic lymphoid leukemia respectively were collect and construction by GraphPad Prism 5.
Verification of Expression for the Differentially Expressed tsRNA
Total RNA was extracted to synthesize cDNA according to the manufacturer’s protocol. In brief, the steps for RNA extraction are as follows: centrifuge to collect 2.5 ×107 cells, transfer a 1.5 mL tube, added 1mLTrizol, set aside at room temperature for 5min; add 0.2mL chloroform, shake for 15s, and let stand for 2min; cen- trifuge at 4â??, 1200g ×15min, and collect the supernatant; add 0.5mL isopropanol, gently mix the liquid in the tube and let stand at room temperature for 10min; centrifuge at 4â??, 12000g ×10min, and the supernatant; add 1mL 75% ethanol, gently wash the pre- cipitate; centrifuge at 4â??, 7500 ×5min, and discard the super- natant; dry and add an appropriate amount of DEPC to dissolve. The RNA samples were QC’d for quantity by NanoDrop ND- 1000 spectrophotometer and RNA integrity by Bioanalyzer 2100 or gel electrophoresis. RT-PCR was performed. U6 was chosen as internal control for tsRNA Quantification in cell samples. The primer as: tRF5-GlyCCC: F:5’ATCGCGCCGCTGGTGTAGT3', R:5’CGTGTGCTCTTCCGATCTTTG3, 5'tiRNA-GlnCTG:F:5’ CGACGATCGGTAGTGTAGTCTACTG3',R:5’ TTCCGATCT- CAGAGCCCAAG3’;5'tiRNA-ArgTCG: :5’TCTACAGTC- CGACGATCGGC3', R:5’ AAGTCAGACGCCTTATCCATT- AG3’; tRF5-TyrGTA: F:5’ CAGTCCGACGATCTCTTCAATA3', R:5’ CTTCCGATCTCGCTCTACCA3’;U6: :5’GCTTCGGCAG- CACATATACTAAAAT3’,R:5’CGCTTCACGAATTTGCGTGT-CAT’. The relative expression levels of tsRNAs were calculated by standard curve method.
Statistical Analysis
The study performed biological replicates. Statistical analysis and statistical figures presented were performed with GraphPad Prism V5.0 software and SPSS22.0 software. t-test was used for comparison between groups. Pâ?»0.05 was statistically significant.
Results
Detection of Resistance for HL-60 Cells
To detect the resistance of HL-60 cells to ADM, we performed cell viability and drug-resistant protein expression. The viability of sensitive and resistant cells were statistically different when the concentration of ADM greater than 0.25 μg/mL. The result of drug- resistant protein expression showed that MDR1 in drug-resistant cells was expressed significantly higher than that in sensitive cells, as shown in Figure 1, indicating that the construction of HL-60/ ADM drug-resistant cell lines were completed.

Figure 1: Detection of HL-60/ADM Resistance A) HL-60/ADM cell viability, the ADM concentration gradient was set at 0, 0.25, 0.5, 1, 2, 4 and 8 μg/mL. The horizontal axis is ADM concentration and vertical axis is cell viability (*P0.25=0.032, **P0.5=0.005, ***P1=0.001, **P2=0.003, ***P4<0.001, ***P8>0.001. B) Expression of drug-resistance protein MDR1. C) MDR1/β-actin ratio (***P=0.001).
Expression of miRNA and tsRNA in Resistant Cells
To analyze genes expression in AML drug-resistant cells, we performed arraystar small RNA microarray analysis on miRNA, Pre-miRNA, tRNA and tsRNA, and the differentially expressed genes were screened which were 337,13,7 and 355, respectively. Volcanic maps were constructed based on fold change and p-value (cut off 0.05), provided in Figures 2A (tRNA), 2B (tsRNA), 2G (miRNA) and 2H (Pre-miRNA). Hierarchical cluster analysis showed that the expression patterns of HL-60/ADM and HL- 60 cells were significantly different (tRNA, Figure 2C; tsRNA, Figure 2D; miRNA, Figure 2I, Pre-miRNA, Figure 2J). Scatter plots were used to evaluate tRNA, (Figure 2E), tsRNA(Figure 2F), miRNA(Figure 2K) and Pre-miRNA(Figure 2L) between HL-60/ ADM and HL-60 cells.
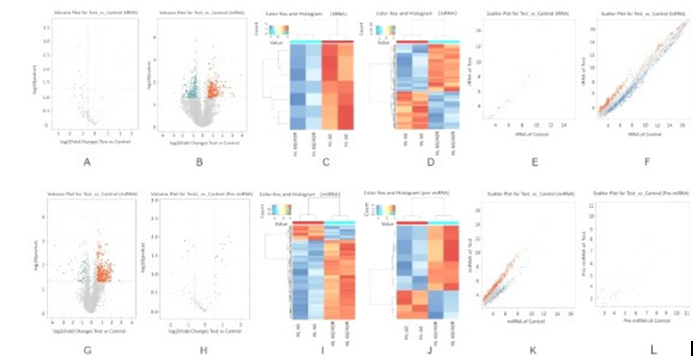
Firgure 2: Differential Expression of miRNA, Pre-miRNA, tRNA and tsRNA in HL-60 and HL-60/ADM. A-F: Volcano Plots, Hierarchical Clustering and Scatter Plots of the Differentially Expressed tRNA and tsRNA in Cell, Respectively, and G-L Which were what miRNA and Pre-miRNA Shown
Characteristics of Differentially Expressed Genes
To comprehend more about the genes screened, we performed characteristic analysis. The results showed that tsRNA aggregated for 49.86%, amount to nearly half in the total differentially expressed genes of miRNA, pre-miRNA, tRNA and tsRNA, suggesting that the dysregulated tsRNA was surprisingly higher than miRNA(Figure 3A). Next, we classified the molecular mechanism of maladjusted tsRNA according to the reported studies. Among them, associated with the development of CLL, accounted for the highest proportion as 73.43%, followed by piRNA-like function, making up 20.41% (Figure 3B). We also carried out statistics of different types of tsRNAs, the proportion of upregulated tsRNAs shown in Figure 3C and 3D are downregulated tsRNAs. Meanwhile, for better understand the possible functions of different types of tsRNAs, we made a schematic diagram of their production which shown in Figure 3E.
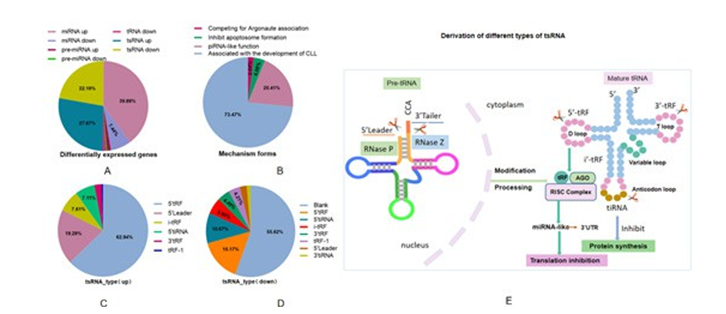
Figure 3: Characteristics of Differentially Expressed Genes A) Differentially expressed miRNAs, pre-miRNAs, tRNAs and tsRNAs proportion pie chart, B) Mechannism forms of dysregulated tsRNAs, C-D) The proportion of differentially expressed tsRNA; E: Schematic diagram of derivation of different types of tsRNAs
Functions of Differentially Expressed-Related tsRNAs
In the purpose of understand the molecular functions of differentially expressed tsRNAs, categorization statistics were performed base on the reported studies. As shown in the Figure 4A, 10 tsRNAs that associated piRNA-like functions. In the associated with the devolepment of CLL module, including 16 up-regulated tsRNAs and 21 down-regulated (Figure 4 B, C).
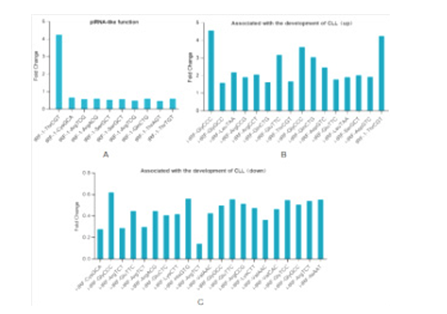
Figure 4: Functions of Differential Gene-Related tsRNAs A) tsRNAs that associated piRNA-like functions, B-C) Up-regulated and down-rugulated tsRNAs which associated with the devolepment of chronic lymphoid leukemia respectively
3.5. Expression Validation of tsRNA by RT-PCR
For validatethe expression of differentially expressed genes in cells, RT-PCR was used to verify the relative expression levels of tsRNAs. Results as shown in Figure 5, tRF5-GlyCCC and 5'tiRNA- GlnCTG were significantly up-regulated, while 5'tiRNA-ArgTCG and tRF5-TyrGTA were significantly down-regulated, which were consistent with the results of arraystar small RNA microarray. These findings confirm that abnormal tsRNA expression patterns occur in AML resistant cells, and may play an important role in the prognosis.
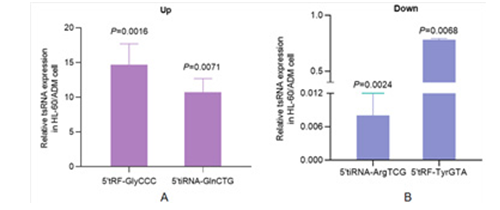
Figure 5: Expression validation of tsRNA by RT-PCR A) Relative expression of up-regulated tsRNA in HL-60/ADM compared with HL- 60 (tRF5-GlyCCC: **P<0.01, 5'tiRNA-GlnCTG: **P<0.01), B) Expression of down-regulated tsRNA (5'tiRNA-ArgTCG: **P<0.01, tRF5-TyrGTA: **P<0.01)
Discussion
Since the last decade, sncRNAs have been increasingly researched in tumor-related studies, and corroborated play an important role in the development of tumorigenesis, metastasis, and drug resistance [6]. Increasing lines of evidences have confirmed that dysregulation of tsRNA could affect various cellular processes such as cell proliferation and invasion in recent years [8]. tsRNA is widely present in prokaryotes and eukaryotes, showing a temporal and spatial expression pattern to play supposed function. tsRNA biogenesis is dynamically regulated during development, the expression is unique to tissues and cells, which can be as a new biomarker [22,23].
AS a new type of small RNA, tsRNA mostly produced by specific nucleases in specific cells or tissues or under certain conditions, such as stress and hypoxia [24]. The expression profile and biological function of tsRNAs have been reported CLL, lung cancer, colorectal cancer, breast cancer and prostate cancer [25- 29]. These studies suggest that tsRNAs may have therapeutic targets and prognostic potential in cancer. However, this has not been reported in AML resistant cells. Therefore, we determined the expression profile of tsRNA in AML resistant cells by using arraystar small RNA microarray analysis.
A total of pre-miRNAs, miRNAs, tRNAs and tsRNAs were detected in AML resistant cells were 170, 2575, 157 and 4067 respectively. Among them, 337 differentially expressed miRNAs were slightly lower than that of tsRNA (345), indicating that dysregulated tsRNAs were highly enriched in AML resistant cells and may even exceed the intracellular miRNAs, indicating that dysregulated tsRNAs play a critical role in the regulation of AML chemotherapy resistance. Subsequently, we analyzed the categories of tsRNA and found that the highest proportion was 5'-tRF, followed by 5'-tiRNA. It has been reported that tRFs and tiRNAs can promote cell proliferation and cell cycle progression by regulating the expression of oncogenes, tRFs can also bind to RNA-binding proteins to inhibit cancer progression, similar to miRNAs, tRFs can inhibit the expression of cancer-related genes, show in argonaute (Ago) protein binds to the 3 'untranslated region (3'UTR) of targeted mRNA to form a RNA-induced silencing complex (RISC) and inhibit oncogene expression at the post- transcriptional or translational level [30].
The dysregulation of tRNA-derived fragments (tRFs) and tRNA- derived stress-induced RNAs (tiRNAs) in AML resistant cells may be caused by abnormal regulation of tRNAs and tiRNAs, and the expression of tsRNAs may be disregulated when oncogenes are activated or suppressor genes are inactivated. Accumulating evidence suggests that some modifications on tRNAs can control the abundance of tsRNA, these modifications enhance tRNA stability and protect tRNA from being cleavage by ANG, as a result the loss of tRNA modifications leads to upregulation of tsRNA [31,32]. In addition, we analyzed the mechanism of the existing records of the differentially expressed tsRNAs and found that many tsRNAs were
associated with CLL. It is suggested that the abnormal expression of tsRNA may also occurs in hematological diseases and plays an important role in the occurrence and progression of the disease.
Dysregulation of tsRNA expression is a concomitant event of tumorigenesis and development. For any up-regulated or down- regulated genes, we are eager to comprehend whether these changes are the initiators of certain malignancies, or the outcome of many genes during malignant transformation. In CLL, the limited available studies have revealed that dysregulation of different types of tRFs is a downstream event, and these fragments may be valuable markers of CLL progression, drug resistance, and/or prognosis [33]. In this study, four tsRNAs with high enrichment were selected for relative expression verification, suggesting that these four dysregulated tsRNAs may be prognostic markers for the occurrence of AML resistance cells. Zhao statemented METTL1 mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control [20]. A related study is Xia who showed that serum tsRNAs to be closely associated with AML prognosis, suggesting the potential application of serum tsRNAs as biomarkers to assist in AML diagnosis [19]. Similar studies have also revealed abnormal expression patterns of AML tsRNA at different levels, but this study is the first to report the screening and identification of tsRNA in HL-60/ADM cells, which provides a theoretical basis for AML drug resistance research in the future.
Overall, this study determined the tsRNA expression profiles of AML drug-resistant cells, provided new insights into the mechanism of HL-60 cells drug resistance, and a theoretical basis for the study of tsRNA. However, it cannot be denied that there are many shortcomings in this study. For example, this study only obtained relevant results at the HL-60 cells which modeling drug resistance solely with Adriamycin (ADM) and lacked validation of animal and clinical samples. Meanwhile, it has not yet studied the function and mechanism of related genes, which is an urgent task for us to solve in the next work.
Conclusion
In conclusion, tsRNA expression profiles in Adriamycin-resistant acute myeloid leukemia cells were screened and identificated in present study, suggesting that tRF5-GlyCCC and 5'tiRNA-GlnCTG may be biomarkers of poor prognosis in AML. Nevertheless, 5'tiRNA-ArgTCG and tRF5-TyrGTA may actively regulate AML to counter chemotherapy resistance. This provides a new insights for the mechanism and therapeutic targets of chemoresistance in AML.
Acknowledgements
This study was supported by Guizhou Provincial Health Commission Science and Technology Plan project (No. gzwkj2023-029 and No. gzwkj2025-047). Meanwhile, we appreciate the help of Kangcheng Biotechnology in Shanghai, China, in the arraystar small RNA microarray analysis.
References
1. Sun, J., Ning, S., Feng, R., Li, J., Wang, T., Xing, B., ... & Liu, H. (2022). Acute myeloid leukemia with cup-like blasts and FLT3-ITD and NPM1 mutations mimics features of acute promyelocytic leukemia: a case of durable remission after sorafenib and low-dose cytarabine. Anti-Cancer Drugs, 33(1), e813-e817.
2. Koreth, J., Schlenk, R., Kopecky, K. J., Honda, S., Sierra, J., Djulbegovic, B. J., ... & Cutler, C. (2009). Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: systematic review and meta-analysis of prospective clinical trials. Jama, 301(22), 2349-2361.
3. Short, N. J., & Kantarjian, H. (2021). When less is more: reevaluating the role of intensive chemotherapy for older adults with acute myeloid leukemia in the modern era. Journal of Clinical Oncology, 39(28), 3104.
4. Coombs, C. C., Tallman, M. S., & Levine, R. L. (2016). Molecular therapy for acute myeloid leukaemia. Nature reviews Clinical oncology, 13(5), 305-318.
5. Döhner, H., Estey, E., Grimwade, D., Amadori, S., Appelbaum, F. R., Büchner, T., ... & Bloomfield, C. D. (2017). Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood, The Journal of the American Society of Hematology, 129(4), 424-447.
6. Wang, T., Cao, L., He, S., Long, K., Wang, X., Yu, H., ...
& Li, W. (2020). Small RNA sequencing reveals a novel tsRNAâ?06018 playing an important role during adipogenic differentiation of hMSCs. Journal of Cellular and Molecular Medicine, 24(21), 12736-12749.
7. Sun, C., Fu, Z., Wang, S., Li, J., Li, Y., Zhang, Y., ... & Yin, Y. (2018). Roles of tRNA-derived fragments in human cancers. Cancer letters, 414, 16-25.
8. Zhou, J., Wan, F., Wang, Y., Long, J., & Zhu, X. (2019). Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer. Cancer management and research, 3945-3956.
9. Jin, F., Yang, L., Wang, W., Yuan, N., Zhan, S., Yang, P., ... & Wang, Y. (2021). A novel class of tsRNA signatures as biomarkers for diagnosis and prognosis of pancreatic cancer. Molecular cancer, 20(1), 95.
10. Xue, M., Shi, M., Xie, J., Zhang, J., Jiang, L., Deng, X., ... & Chen, H. (2021). Serum tRNA-derived small RNAs as potential novel diagnostic biomarkers for pancreatic ductal adenocarcinoma. American journal of cancer research, 11(3), 837.
11. Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., ... & Peng, Y. (2019). Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Molecular cancer, 18, 1-5.
12. Zhao, C., Tolkach, Y., Schmidt, D., Kristiansen, G., Müller, S. C., & Ellinger, J. (2018). 5′-tRNA halves are dysregulated in clear cell renal cell carcinoma. Journal of Urology, 199(2), 378-383.
13. Wang, B., Xia, L., Zhu, D., Zeng, H., Wei, B., Lu, L., ... & Sun, M. (2022). Paternal high-fat diet altered sperm 5'tsRNA- Gly-GCC is associated with enhanced gluconeogenesis in the offspring. Frontiers in molecular biosciences, 9, 857875.
14. Karousi, P., Adamopoulos, P. G., Papageorgiou, S. G., Pappa, V., Scorilas, A., & Kontos, C. K. (2020). A novel, mitochondrial, internal tRNA-derived RNA fragment possesses clinical utility as a molecular prognostic biomarker in chronic lymphocytic leukemia. Clinical Biochemistry, 85, 20-26.
15. Katsaraki, K., Artemaki, P. I., Papageorgiou, S. G., Pappa, V., Scorilas, A., & Kontos, C. K. (2019). Identification of a novel, internal tRNA-derived RNA fragment as a new prognostic and screening biomarker in chronic lymphocytic leukemia, using an innovative quantitative real-time PCR assay. Leukemia Research, 87, 106234.
16. Balatti, V., Pekarsky, Y., & Croce, C. M. (2017). Role of the tRNA-derived small RNAs in cancer: new potential biomarkers and target for therapy. Advances in cancer research, 135, 173-187.
17. Hu, F., Niu, Y., Mao, X., Cui, J., Wu, X., Simone, C. B., ... & Jiang, L. (2021). tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Translational lung cancer research, 10(10), 3957.
18. Balatti, V., Nigita, G., Veneziano, D., Drusco, A., Stein, G. S., Messier, T. L., ... & Croce, C. M. (2017). tsRNA signatures in cancer. Proceedings of the National Academy of Sciences, 114(30), 8071-8076.
19. Xia, L., Guo, H., Wu, X., Xu, Y., Zhao, P., Yan, B., ... & Zhang, X. (2023). Human circulating small non-coding RNA signature as a non-invasive biomarker in clinical diagnosis of acute myeloid leukaemia. Theranostics, 13(4), 1289.
20. Zhao, P., Xia, L., Chen, D., Xu, W., Guo, H., Xu, Y., ... & Zhang, X. (2024). METTL1 mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control. Experimental Hematology & Oncology, 13(1), 8.
21. Li, M., Meng, F., & Lu, Q. (2020). Expression profile screening and bioinformatics analysis of circRNA, LncRNA, and mRNA in acute myeloid leukemia drug-resistant cells. Turkish Journal of Hematology, 37(2), 104-110.
22. Li, J., Zhu, L., Cheng, J., & Peng, Y. (2021, October). Transfer RNA-derived small RNA: a rising star in oncology. In Seminars in cancer biology (Vol. 75, pp. 29-37). Academic Press.
23. Zhu, L., Liu, X., Pu, W., & Peng, Y. (2018). tRNA-derived small non-coding RNAs in human disease. Cancer letters, 419, 1-7.
24. Shen, Y., Yu, X., Zhu, L., Li, T., Yan, Z., & Guo, J. (2018). Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. Journal of Molecular Medicine, 96, 1167-1176.
25. Katsaraki, K.,Adamopoulos, P. G., Papageorgiou, S. G., Pappa, V., Scorilas, A., & Kontos, C. K. (2021). A 3′ tRNAâ?derived fragment produced by tRNALeuAAG and tRNALeuTAG is associated with poor prognosis in Bâ?cell chronic lymphocytic leukemia, independently of classical prognostic factors. European Journal of Haematology, 106(6), 821-830.
26. Shao, Y., Sun, Q., Liu, X., Wang, P., Wu, R., & Ma, Z. (2017). tRFâ?Leuâ?CAG promotes cell proliferation and cell cycle in nonâ?small cell lung cancer. Chemical biology & drug design, 90(5), 730-738.
27. Mo, D., Jiang, P., Yang, Y., Mao, X., Tan, X., Tang, X., ... & Yan, F. (2019). A tRNA fragment, 5′-tiRNAVal, suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer letters, 457, 60-73.
28. Zhang, M., Li, F., Wang, J., He, W., Li, Y., Li, H., ... & Cao, Y. (2019). tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. OncoTargets and therapy, 6371-6383.
29. Yang, C., Lee, M., Song, G., & Lim, W. (2021). tRNALys-derived fragment alleviates cisplatin-induced apoptosis in prostate cancer cells. Pharmaceutics, 13(1), 55-61.
30. Zhu, L., Ge, J., Li, T., Shen, Y., & Guo, J. (2019). tRNA- derived fragments and tRNA halves: the new players in cancers. Cancer Letters, 452, 31-37.
31. Wang, X., Matuszek, Z., Huang, Y., Parisien, M., Dai, Q., Clark, W., ... & Pan, T. (2018). Queuosine modification protects cognate tRNAs against ribonuclease cleavage. Rna, 24(10), 1305-1313.
32. Chen, Z., Qi, M., Shen, B., Luo, G., Wu, Y., Li, J., ... & Wang, H. (2019). Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic acids research, 47(5), 2533-2545.
33. Veneziano, D., Tomasello, L., Balatti, V., Palamarchuk, A., Rassenti, L. Z., Kipps, T. J., ... & Croce, C. M. (2019). Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences, 116(48), 24252-24258.